PhD
Assistant Professor
UNC-Chapel Hill
Cancer Cell Biology
Area of Interest
Our work is focused on signal transduction in single human cells. We are specifically interested in why genetically identical cells choose different fates and how these decisions are influenced by the cellular environment. Our lab uses a variety of experimental and theoretical approaches including transcriptional profiling, time-lapse microscopy, and computational modeling. Like many basic research labs, we work under the fundamental assumption that if we gain a mechanistic understanding of how cells process molecular signals, we can apply this knowledge to design therapies for cancer and regenerative medicine.
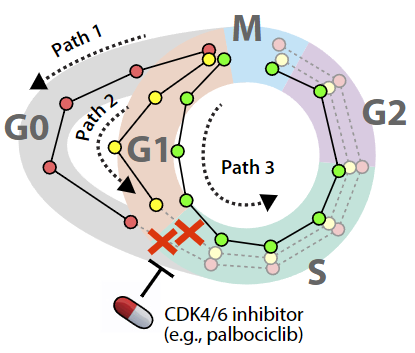
A major question in the lab is how cells enter senescence—a state of permanent cell cycle arrest. Senescence plays a role in tumor suppression, aging, and differentiation, but its precise involvement in these processes is poorly understood. For example, senescence has been shown to stop the formation of tumors by shutting down the cell cycle in premalignant cells. Yet it also appears to promote tumor growth in some contexts and has been linked to aging, embryonic development, and inflammation. Mutations or deletions of key senescence genes such as TP53 (p53) and CDKN2A (p16INK4a and p14ARF) are extremely common in human cancers and attest to the importance of this pathway in disease etiology. Surprisingly, many fundamental questions about senescence remain to be answered: When do cells commit to senescence? Is senescence truly irreversible? How, mechanistically, do stress signals trigger expression of senescence genes? Addressing these questions will yield valuable knowledge for basic cell biology and promote new strategies to induce senescence in tumor cells. We are using state-of-the-art experimental and computational methods to study senescence in individual cells. Using live-cell fluorescent imaging, we are able to monitor cell divisions, key senescence markers, and upstream stress signals in real time and at single-cell resolution. Our overarching hypothesis is that—by identifying the precise time at which individual cells commit to permanent arrest—we can pinpoint the molecular mechanisms that govern this decision. This knowledge could lead to new strategies for inducing senescence in tumor cells.
Another major question we are studying is how signaling dynamics affect cell fate decisions. That is, how do the temporal patterns of a signaling molecule control downstream responses? After finding that the dynamics of the tumor suppressor p53 regulate gene expression and cell fate, we suspect that many other signaling pathways behave in a similar way. We are currently focused on the p38 MAP kinase, which, like p53, responds to various cellular stresses and can trigger multiple cellular outcomes. Previous studies have shown that transient p38 activation is associated with cell proliferation, while sustained p38 activity leads to cell cycle arrest. To directly test this hypothesis, we are using a synthetic p38 kinase to directly control its activity in living cells. We use a variety of approaches to assess the downstream response of individual cells such as live-cell reporters and single-molecular fluorescent in situ hybridization. In all of our work, we use computational models to help describe, quantify and predict cellular responses.
In summary, we use modern single-cell and computational approaches to understand how cells interpret normal and pathological signals. We apply these approaches to study major tumor suppressor pathways such as p53, p16INK4a, and the p38 MAP kinase. When possible, we look for opportunities to translate these basic discoveries into molecular therapies that will improve patient health and quality of life.
Awards and Honors
- NIH Pathway to Independence Award – NIGMS (K99/R00), 2012
- Ruth L. Kirschstein National Research Service Award – NIGMS (F32), 2010-2012
- Visiting Scholar at Duke Center for Systems Biology, 2011
- NHGRI Computational Genomics Graduate Fellowship, 2008-2009
- Computational Molecular Science and Engineering Forum (CoMSEF) Graduate Student Award, 2008
- James Davidson Graduate Travel Scholarship, 2004
- American Society for Microbiology Travel Award, 2004
- President’s Award, American Society for Microbiology, 2003
- Robert C. Byrd Honors Scholarship, 1997-2001
- University of Florida Honors Program, 1997-1999