Our Research
Overview
Our laboratory is unique in being able to combine sophisticated biochemistry, genetics, and molecular biology with the ability to directly visualize DNA, proteins, and DNA or RNA-protein complexes, as well as whole cells using high resolution electron microscopes. Most of the methods for preparing the samples are ones we developed and thus we have an expertise which few other groups possess. This has resulted in many research papers of significance, including the first visualization of nucleosomes, the first visualization of bent DNA, and the discovery of telomere looping. We have a wealth of collaborations with laboratories world-wide which greatly enrich our research program and the training environment for laboratory members who frequently collaborate and publish with top groups elsewhere. I keep the lab relatively small so that I too can carry out experiments at the bench, which I have continued to do my whole career. All members of the laboratory, graduate students, fellows, and technicians are considered equals, and several years ago the whole group traveled to Alaska in the summer for a workshop on aging and DNA repair we organized.
T-loops and R-loops
R-loops: a new form of DNA damage at the telomere
Telomeres contain ~1/3000th the DNA in a human cell, yet shelter us from oncogenic progression, monitor the number of cell divisions and respond to toxic insult from agents producing DNA damage. This is accomplished via pathways which signal the presence of DNA lesions. A central feature of the telomere is its 3-dimensional structure, and telomeres are rich in complex structural elements including the t-loop, t-loop junction (more complex than previously believed), and R-loops. The latter are a recently recognized component of telomeres. R-loops can lead to genomic instability, gross chromosomal rearrangements, and cancer. R-loops may represent one of the greatest threats to telomeres yet may be needed to form protective t-loops.
Studies of R-loops formed in genomic and telomeric DNA has led to an appreciation of these 3-stranded structures in both maintaining and damaging the genome. R-loops can shelter promoters from DNA methylases promoting transcription of over 1220 human genes. At a stalled fork, the displaced ssDNA in an R-loop can be cleaved resulting in DSB’s, unwanted recombination, and genomic instability. R-loops are more prevalent in BRCA2 and topo I deficient cells and can be several hundred bp in size. R-loop formation has been implicated in ALS type 4 and Fragile X diseases.
R-loops at the telomere (tel-R-loops) are more stable due to the formation of G quadruplexes on the displaced strand. Azzalin et al showed that depleting RNase H1 results in more tel-R-loops, replication stress, telomere shortening dependent on RPA and generation of t-circles. They showed that tel-R-loops are tightly regulated by RNase H1,2 levels which remove them, and loss of these nucleases result in elevated TERRA RNA (G-rich telomeric RNA transcript) and tel-R-loops resulting in the expression of the ALT phenotype (Alternative Lengthening of Telomeres). A dramatic case of tel-R-loops is the ICF syndrome (Immunodeficiency, centromeric instability and facial anomalies syndrome). Cells from patients show abnormally high levels of TERRA (100 fold) and high levels of tel-R-loops. This is due to the hypomethylated state of the TERRA promoters from a biallelic loss of DNMT3B methylase.
Tel-R-loops may help form t-loops
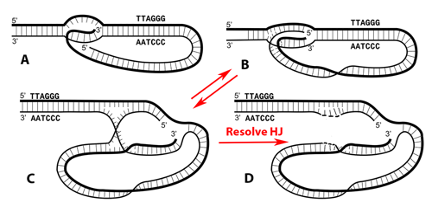
In 1999 we showed that the ends of mammalian telomeres loop back and invade the preceding telomeric DNA. T-loops have now been found from humans to yeast (with genetic manipulation). Recently Doksani in the de Lange and Zhang labs used STORM light microscope imaging to confirm the presence of t-loops in mouse cells and observed loop frequencies and dimensions similar to what we observed by EM. Knockdown of TRF2 resulted in a 5-fold reduction in t-loops.
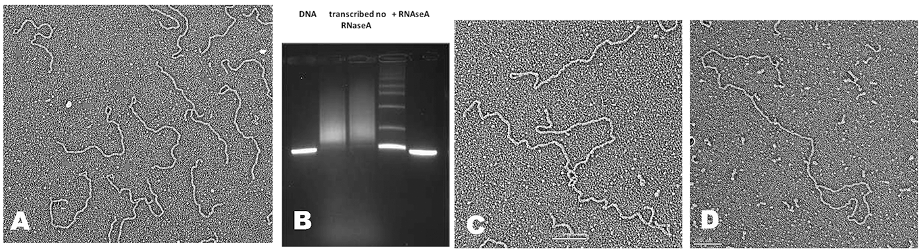
While this seems to point to TRF2 creating loops, the STORM studies only showed that TRF2 is needed to maintain them in vivo and it was unclear what events in the cell generate t-loops. Cesare and colleagues have confirmed and extended the Doksani study. Clues into what drives loop formation came from the discovery of telomeric RNA. Telomeres are transcribed by RNA polymerase II from sub-telomeric promoters. The C-rich strand is predominantly transcribed generating a G-rich RNA termed TERRA. TERRA transcripts range from short to 9000 nt. TERRA is accepted as a key structural element of telomeres, providing a scaffold upon which TRF1 and TRF2, hnRNPA1 and Orc1 will bind. TERRA peaks in G1, declines through S phase and rises in G2. TERRA levels are increased in human cancer cells and in ALT. In 2016 we asked whether transcription of TERRA in a model telomere system might facilitate t-loop formation by opening the helix and leaving behind tel-R-loops and also promote homologous recombination (HR), two features of tel-R-loops in vivo. The telomere model DNA employs our plasmid containing a T7 promoter followed by 576 bp of TTAGGG repeats. Its transcription generates TERRA and we discovered drives a high fraction of the DNA into t-loops (>50%) and also HR products as seen by EM and agarose gel electrophoresis (Fig 1B below). Loop formation did not require a single strand (ss) overhang and thus both strands at the terminus of the telomeric tract must be inserted back into the preceding duplex (Fig. 2). The t-loops generated this way are extremely stable, and more so when TERRA remains present. The t-loop junctions frequently contain an RNase resistant bead containing TERRA and thus represent very complex tel-R-loops. We developed a method to generate pure telomeric DNA, either ss or double strand (ds) up to 15,000 nt (or bp) in length beginning with a T7 promoter. Transcription of the human sized dsDNA generated t-loop molecules with long tails and circular portions as large as 5 kb, typical of human t-loops cells (Fig. 1D).
These studies are described in several of our recent publications and summarized in the review in DNA repair by Tomaska et al (2020). They are providing a new platform for thinking about telomeres and further work designed to probe the models presented.
G-quadruplex induced condensation of G-rich ssDNA
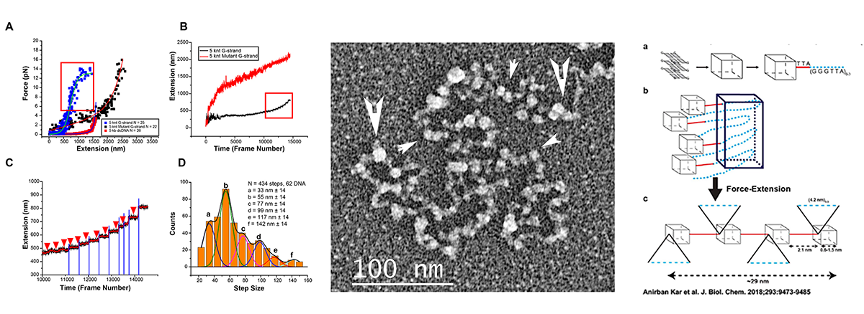
We recently developed a rolling circle replication method for generating very high molecular weight ssDNA of any repeating sequence. This novel technology made it possible to generate single stranded or double stranded DNA the size of human telomeres containing a T7 RNA polymerase promoter at the beginning. This allowed us to ask what structures form on long ssDNA consisting of (TTAGG)n repeats as well as the C-strand (AATCCC)n or mutants thereof. Our EM studies showed that the G-rich ssDNA self-condenses into particles 8-10 nm in diameter (EM, Fig. 3 center below, arrows indicate large and small beads). Analysis of size-selected ssDNA showed that each bead contains ~120 or 240 nt. Fluorescence displacement assays revealed the formation of G-quadruplexes in the G-rich ssDNA. Single molecule magnetic tweezer (smMT) force extension analysis carried out with the laboratory of Richard Fishel at Ohio State University (Fig. 3A-D) of 5000 nt ssDNAs showed a stepwise extension [B,C,D] for the G-rich ssDNA but not the C-rich or mutant (TTAGCG)n ssDNAs (A,B). Fig. 3A shows a model (far right) in which particles are formed as flat disks consisting of a 24 nt G stabilized by quadruplexes as based on X ray studies. New studies will be focused on a full cryoEM structural study of these large particles formed on the G-rich ssDNA.