Research Domains
As we move forward with activities related to sequence-based diagnostics in LMIC, Megan Roberts, PhD, (UNC Eshelman School of Pharmacy) is leading a study to develop an implementation framework for genomic sequencing as a diagnostic test for pediatric cancer in low- and middle-income countries. Implementation science is needed to develop approaches, resources, and processes to overcome identified barriers. Implementation science is the study of methods and strategies that facilitate the integration of evidence-based practice and research into clinical use.

This field is particularly important for the implementation of complex interventions, such as sequencing-based diagnostics which has multiple interdependent components including implementation of the technology, return of results, and de-implementation of traditional diagnostic technologies. Using frameworks and methods from the field of implementation science, we are (1) developing and testing a multicomponent implementation strategy for sequencing-based diagnostics in pediatric cancer; (2) co-developing a format for return of sequencing-based diagnostic results to providers practicing in diverse pediatric cancer care settings; and (3) developing a process for de-implementation of traditional pathologic tests.
To form a preliminary understanding of the implementation barriers and facilitators for sequencing-based diagnostics, we have already performed 39 semi-structured interviews across 6 pediatric cancer centers. In addition to describing major barriers and facilitators for implementation interviewees offered potential strategies to address implementation barriers including recommending changes to infrastructure, utilization of financial strategies, additional clinician supports (e.g., training and educating implementers about sequencing techniques and interpretation of results; providing interactive assistance), developing relationships, adapting and tailoring the intervention to the context, and using evaluative and integrative strategies to implement sequencing-based diagnostics. This data will inform future strategy development
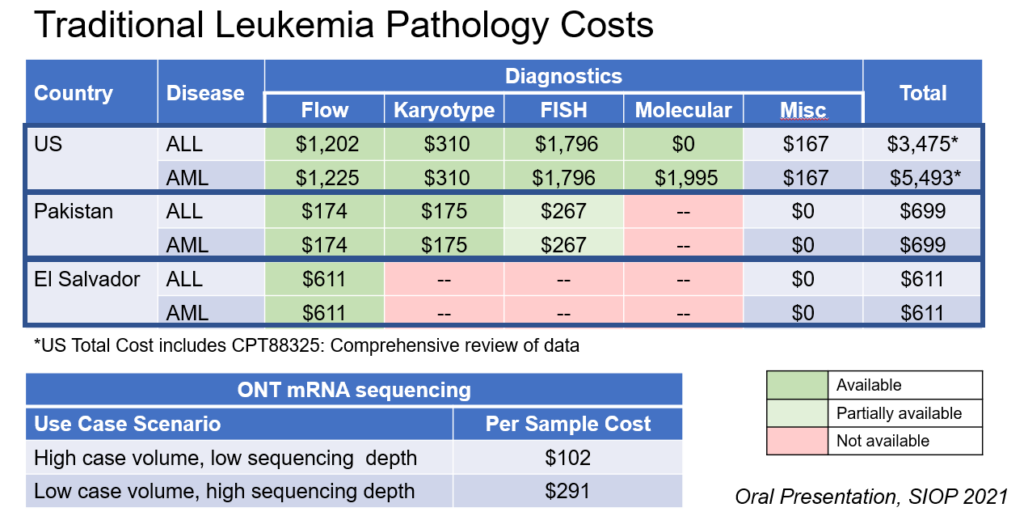 Modern diagnostic methods such as flow cytometry, cytogenetics, and molecular genomics, are standard of care in HICs, but expensive to implement and logistically challenging to maintain in LMICs. To date, the paradigm of building diagnostic capacity in LMIC has followed historical precedent in HICs, with iterative implementation of increasingly complex diagnostic modalities. The feasibility of this approach, both from a cost and logistical perspective, has not been comprehensively studied in LMIC.
Modern diagnostic methods such as flow cytometry, cytogenetics, and molecular genomics, are standard of care in HICs, but expensive to implement and logistically challenging to maintain in LMICs. To date, the paradigm of building diagnostic capacity in LMIC has followed historical precedent in HICs, with iterative implementation of increasingly complex diagnostic modalities. The feasibility of this approach, both from a cost and logistical perspective, has not been comprehensively studied in LMIC.
Moreover, while several publicly accessible databases address medication availability and/or cost in different countries, little to no reliable data have been published about diagnostics, especially in LMIC. The goal of this research component, led by Sachiko Ozawa, PhD (Eshelman School of Pharmacy) and Nickhill Bhakta (St. Jude Children’s Research Hospital) is to evaluate the cumulative costs of current standard diagnostic approaches of pediatric cancer subtypes in LMICs, as a basis for assessing the cost-effectiveness of novel diagnostic methods in these settings.
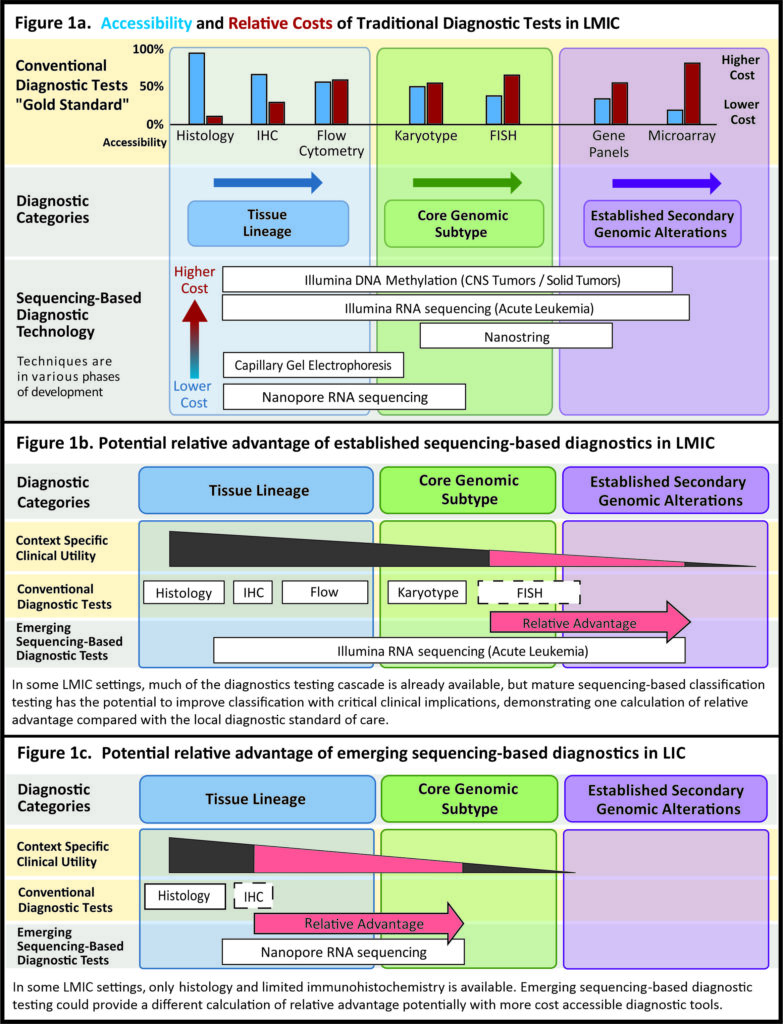 Stuart Rennie, PhD, (UNC Center for Bioethics) is leading members of the bioethics research group to assess ethical outcomes such as beneficence, nonmaleficence, autonomy, and justice, while identifying determinants and strategies to address health inequities related to LMIC implementation of genomic testing for clinical diagnostics. Practically, this work is essential to build trust and consensus among HIC and LMIC clinicians/policymakers that the various ethical trade-offs in molecular approaches are evaluated and shared in a transparent manner. We recently completed a perspective exploration of the concept of relative advantage in sequencing based diagnostics in LMIC (shown in the figure to the right).
Stuart Rennie, PhD, (UNC Center for Bioethics) is leading members of the bioethics research group to assess ethical outcomes such as beneficence, nonmaleficence, autonomy, and justice, while identifying determinants and strategies to address health inequities related to LMIC implementation of genomic testing for clinical diagnostics. Practically, this work is essential to build trust and consensus among HIC and LMIC clinicians/policymakers that the various ethical trade-offs in molecular approaches are evaluated and shared in a transparent manner. We recently completed a perspective exploration of the concept of relative advantage in sequencing based diagnostics in LMIC (shown in the figure to the right).
Moving forward, with a multi-institutional, international group of collaborator, our initial objectives are to engage with collaborators in the context of use to identify potential ethical concerns about sequencing based diagnostics, (b) explore the challenges with return of clinical sequencing-based diagnostic results to providers practicing in diverse pediatric cancer care settings, and (c) develop an ethical framework for de-implementation (or omission of implementation) of traditional pathologic tests (karyotype, fluorescence in site hybridization, and target panels).
 To substantively close the diagnostic gap in pediatric oncology in LMIC, we are adapting affordable genome sequencing for diagnostic classification of pediatric leukemia, lymphoma, and solid tumors.
To substantively close the diagnostic gap in pediatric oncology in LMIC, we are adapting affordable genome sequencing for diagnostic classification of pediatric leukemia, lymphoma, and solid tumors.
Led by Jeremy Wang, PhD, we have developed a sequencing assay for childhood cancer diagnostics from Oxford Nanopore Technologies (ONT, “Nanopore sequencing”), a low-cost sequencing platform. Nanopore sequencing is an established technology with an existing global commercial reach in agriculture and health. As a potential diagnostic tool for childhood cancers, Nanopore sequencing represents a resource-appropriate solution. The platform requires minimal capital investment, limited lab infrastructure and space.
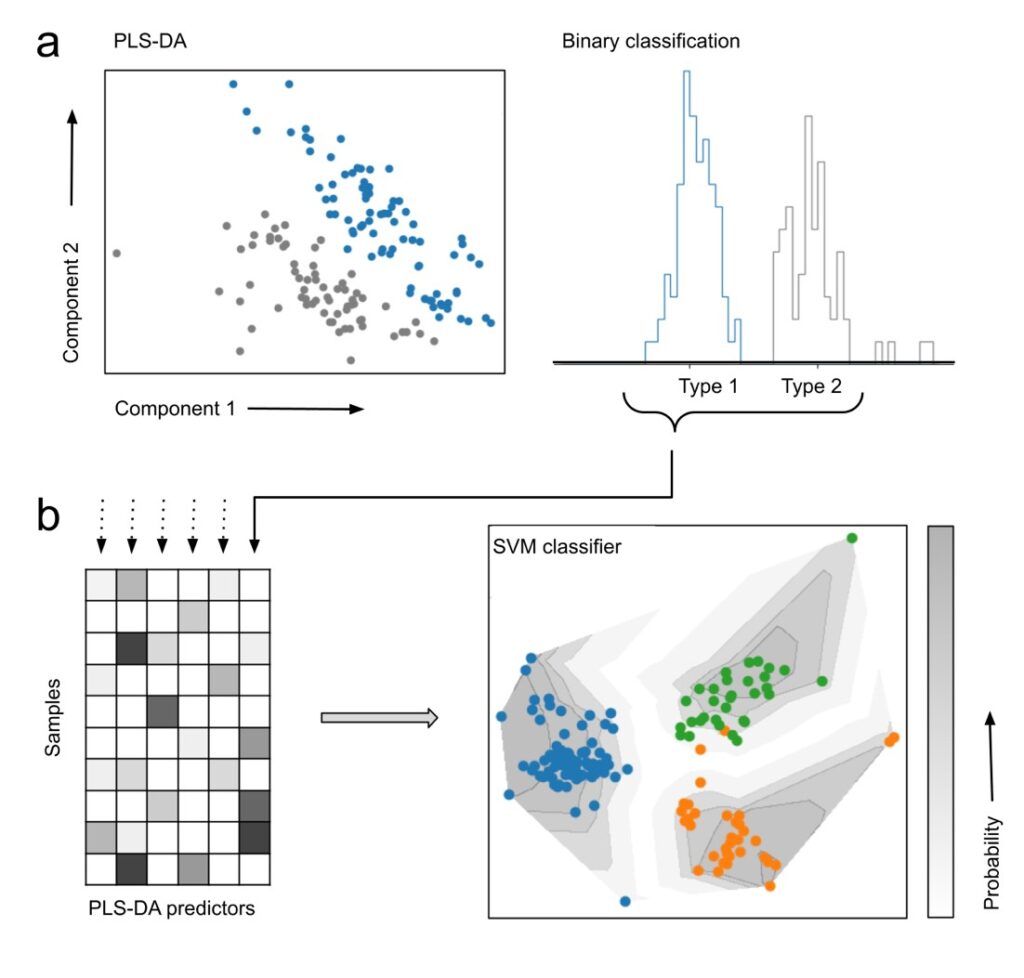
A schematic of our current algorithm, using an approach of partial least squares discriminant analysis, combined with support vector matrix classification, is shown on the right.
We are partnering with Oxford Nanopore Technologies to continue to develop a resource-appropriate technology that will be financially accessible, scalable, and cost-effective across various settings, with immediate and direct clinical implications in LMIC. The cost of Nanopore’s MinION sequencer is less than $1,000 USD, a critical advantage to ensure affordability.
Our approach is innovative by establishing:
- Near-term — resource-appropriate approach to address urgent diagnostic needs in pediatric cancer for the large number of settings without access to comprehensive, consistent immunohistochemistry
- Intermediate-term — advanced classification options for settings with mature risk-adapted treatment strategies by providing genomic sub-types
- Long-term — realistic platform to achieve scalable advancements that cover the diagnostic needs for most childhood cancers through classification and molecular subtype classification