FAQ
Frequently Asked Questions (FAQs)
The Translational Genomics Lab (TGL) is happy to answer any questions you have about our service offerings and your project, even if you are not planning to use us for those services. Below is a list of some frequently asked questions.
General
- Do only Lineberger members have access to TGL services?
- TGL services are subsidized by UNC Lineberger University Cancer Research Funds (UCRF), and our mission is to support translational human cancer genomics, so although we do support projects from non-Lineberger investigators, those projects tend to be directly cancer related. Our priority is to support Lineberger members first, non-Lineberger member with cancer related projects second, and everything else third or not at all. Almost all projects we support fall into the first two categories.
- How can I start a project?
- The first step to any new project is for us to have a consultation to discuss the details of your project. To submit a consultation request, please email ogr@unc.edu
- Do I need to order any reagents or consumables when submitting samples for processing?
- TGL service offering rates include the costs for reagents and consumables, so you do not need to order anything when submitting samples. There are a few exceptions to this rule, but those will be discussed in more detail during the consultation.
- What is the anticipated turnaround time for my project?
- The turnaround time for a project will depend on the number of services being used and the size of the project. During the consultation, an estimated delivery date will be discussed and included in the statement of work (SOW). Below are the average historic turnaround times for projects of any size (a few to thousands of samples). Please keep in mind that smaller projects would likely have shorter turnaround times than those listed below, and larger projects would have longer turnaround times. For projects with impending deadlines (e.g., preliminary data for grants, manuscript revisions), we can expedite processing as needed. Those are details that we discuss during the consultation.
Historic Average Turnaround Times (All Project Sizes Combined)
| Service | Time |
|---|---|
| Nucleic Acid Extraction | 2.4 work weeks |
| Library Preparation and Sequencing | 3.6 work weeks |
| Nucleic Acid Extraction, Library Preparation and Sequencing | 6.3 work weeks |
| Sequencing | 1.2 work weeks |
| NanoString | 2.2 work weeks |
Scientific
- How much DNA and/or RNA yield will I get from my FFPE samples? Can we submit less than 5×10 µm or 10×5 µm scrolls/slides?
- It is very difficult to predict extraction yields from FFPE samples simply based on the thickness of the sections since the yield will depend on the tissue type, total tissue area, and other variables. Our recommendation is to start with the maximum input amount for the kit being used as this will ensure maximum DNA and/or RNA yield, which in general will significantly improve downstream genomic assay success rates.
- How do the extraction yields compare when extracting either DNA or RNA alone versus dual DNA and RNA extraction?
- The dual DNA and RNA extraction workflow first extracts DNA and then RNA, so the DNA yields between the single extraction method and the dual extraction method are the same for DNA. When comparing the extraction yields for RNA, we see only about a 10% lower yield for RNA extracted using the dual extraction workflow compared to the single extraction workflow from the same sample, there does not appear to be a significant decrease in our RNA yields when using the dual extraction method.
- What are the target regions for the UNCseqTM panel?
- For a complete list of current UNCseqTM panel targets, please reach out to lccctgl@unc.edu.
- What are your historic success rates?
- While the assay kits we use are robust, not every sample is expected to pass each step of the genomic workflow. Historic TGL success rates starting from snap frozen and FFPE material are highlighted in the table below. Snap frozen RNA success rates were based on mRNAseq criteria and FFPE RNA success rates were based on total RNAseq criteria. Each set of samples is unique, so the success rates for your project might different than the those listed below.
| Nucleic Acid | Starting Material | Success Rate |
|---|---|---|
| DNA | Snap Frozen | 99.1% |
| DNA | FFPE | 88.9% |
| RNA | Snap Frozen | 86.9% |
| RNA | FFPE | 95.4% |
| Nucleic Acid | Starting Material | Success Rate |
|---|---|---|
| DNA | Snap Frozen | 98.2% |
| DNA | FFPE | 92.9% |
| RNA | Snap Frozen | 98.7% |
| RNA | FFPE | 91.3% |
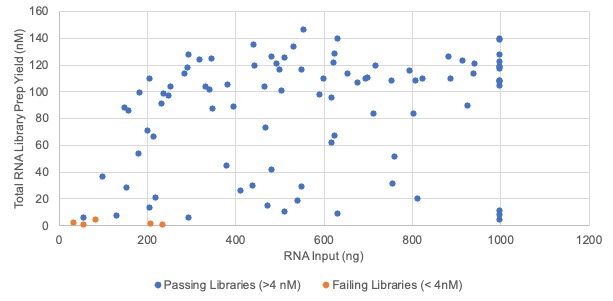
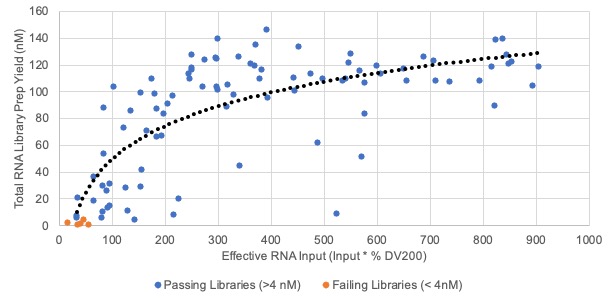
In general, the success rate for genomic assays comes down to how much material you have and the quality. Below is an example of some Illumina TruSeq Stranded Total RNA Library Preps that highlights the importance of looking beyond just quantity and incorporating the quality of the material as well. The top graph shows the library prep yields as a function of RNA Input, while the lower graph shows the library prep yields as a function of Effective RNA Input (Input * %DV200). You can see that incorporating the quality of the RNA into the graph does a better job highlighting the RNA samples most likely to fail the library prep. There is a sharp drop in library prep yields when the Effective RNA Input is less than ~150 ng. This is why maximizing extraction yield and library prep input is essential to these studies since the quality of the FFPE material is something that we have little control over by the time the samples make their way to TGL.
Financial
- My funds are expiring soon. Can you invoice my account before you complete the work?
- UNC School of Medicine Core Facilities are overseen by the Office of Sponsored Programs (OSP) to ensure UNC’s compliance with federal guidelines. While we would like to be able to pre-charge for our services, sponsored projects are subject to Uniform Guidance and University terms and conditions. Uniform Guidance 2 CFR 200.403(h) states that “cost must be incurred during the approved budget period.” Therefore, charging to a project an expense that will not be incurred until after the project ends is not allowable. Furthermore, 2 CFE 200.405(a) states: “a cost is allocable to a particular Federal award or other cost objective if the goods or services involved are chargeable or assignable to that Federal award or cost objective in accordance with relative benefits received.” Costs for services that have not yet been rendered do not benefit the project and are therefore not allowable.
- How do I pay for TGL services?
- TGL services are invoiced through iLab. Since all the details of the project are discussed ahead of time during the consultation and a chart field string (CFS) or PO# is provided with the signed statement of work (SOW), the investigator does not need to directly interact with iLab for services provided by TGL. To simplify the process for our users, it is our standard practice to put all that information into iLab. When an invoice is generated, the primary investigator of the CFS or PO will be emailed a copy of the invoice through the iLab system. For internal users, the invoice is for record keeping purposes only and the CFS provided during the consultation will be automatically charged within a few weeks of the invoice being generated.
- Do Lineberger members receive a discount for TGL services?
- TGL services are subsidized by UNC Lineberger University Cancer Research Funds (UCRF). These discounted service offering rates are the same for all UNC investigators, regardless of Lineberger member status.
To learn more about TGL, email lccctgl@unc.edu or come by 5118A Marsico Hall.
TGL collaborates with the Office of Genomics Research for project management. To submit a consultation request, email ogr@unc.edu