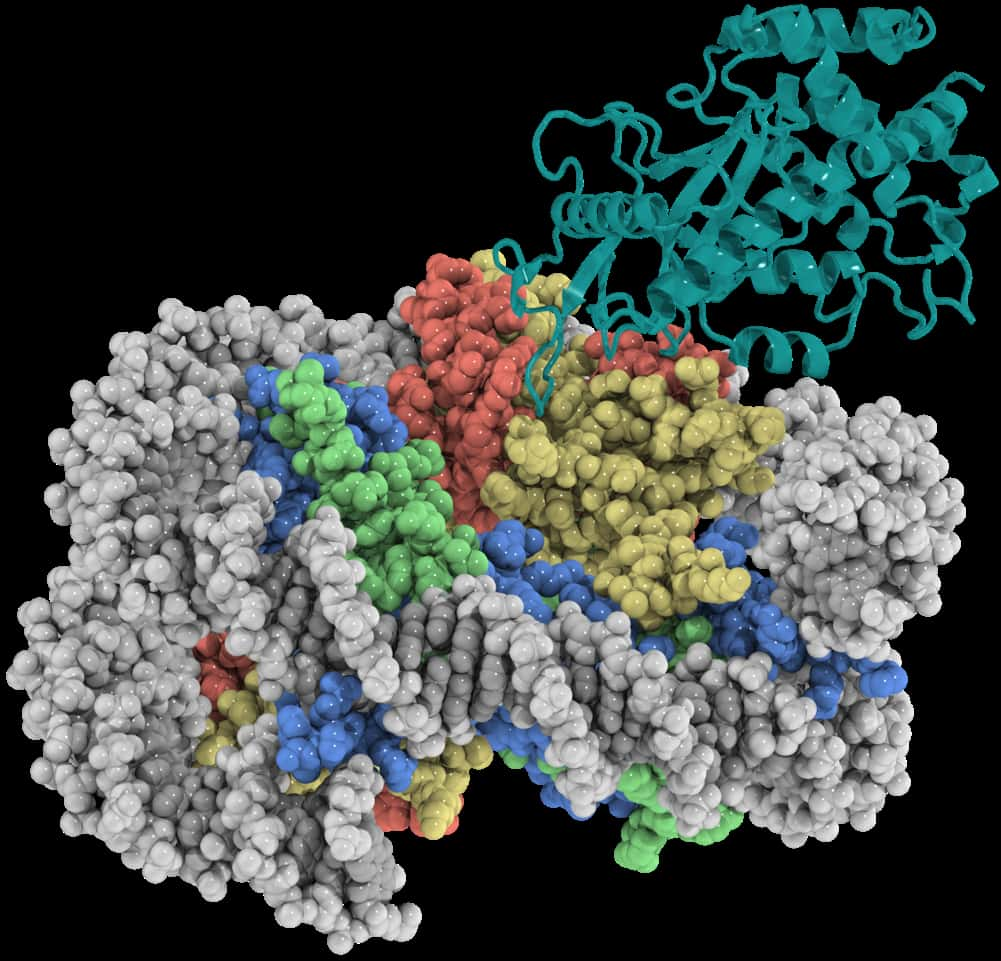
UNC-Chapel Hill researchers have, for the first time, determined the high-resolution structure of a key DNA-sensing protein in the innate immune system called cGAS while it is bound to the nucleosome – the all-important unit of DNA packaging inside a cell’s nucleus.
This research, published in Science, reveals in detail how the nucleosomes inside our cells block cGAS from unintentionally triggering the body’s innate immune response to our own DNA. The work was led by Qi Zhang, PhD, associate professor of biochemistry and biophysics at the UNC School of Medicine, and Robert McGinty, MD, PhD, assistant professor of chemical biology and medicinal chemistry at the UNC Eshelman School of Pharmacy. Both are members of the UNC Lineberger Comprehensive Cancer Center.
“Detecting and responding to foreign DNA from bacterial and viral pathogens is one of the most fundamental mechanisms for host defense,” said Zhang, co-senior author. “A deeper understanding of functions and regulations of this important DNA sensor will have profound impacts on both basic research and translational development of cGAS-targeted therapeutics crucial to the betterment of human health.”
McGinty, co-senior author, said, “This work was enabled by recent advances in cryo-electron microscopy technology that allows scientists, like those on our team, to observe the protein machines inside our cells with unprecedented clarity. By seeing how these proteins function normally, we can gain insights into how to manipulate their functions to treat diseases.”
In the mammalian innate immune system, the protein cyclic GMP–AMP synthase (cGAS) detects foreign or damaged “self” DNAs. Upon DNA detection, cGAS synthesizes cyclic GMP–AMP (cGAMP), the second messenger molecule that activates the cGAS-STING signaling pathway to fight infections, inflammatory diseases, and cancers.
Because cGAS is a “universal” DNA sensor, it must be regulated to differentiate pathogenic DNA from the body’s own healthy DNA to avoid any unintended immune responses. Previous research has shown that cGAS is enriched inside the nucleus where our genomic DNA is stored, but it remains a mystery as how cGAS ignores our own healthy DNA.
Using the UNC School of Medicine state-of-the-art Cryo-Electron Microscopy Core Facility, which was established in 2019, the Zhang and McGinty labs determined a 3.3Å-resolution cryo-EM structure of cGAS in complex with the nucleosome. The structure shows that cGAS employs two conserved amino acids to anchor to a negatively charged patch on the nucleosome surface. These protein-protein interactions allow the nucleosome to occupy a critical DNA sensing surface on cGAS and prevent cGAS from entering its functionally active DNA-bound state. Together with mutagenesis and functional assays, this study provides a near-atomic resolution depiction of how cGAS maintains the resting, inhibited state in the nucleus.
“These findings reshape the current paradigm of cGAS regulation and exemplify the role of the nucleosome in regulating diverse protein functions,” said McGinty, who holds a joint faculty appointment at the UNC School of Medicine.
Zhang added, “Biomedical scientists will be able to apply our research to fields such as immunology, cancer biology, and gene regulation, as well as to drug discovery for infections, inflammatory diseases, and cancers.”
Authors and Disclosures
First authors of the Science paper are Joshua Boyer, PhD, a postdoctoral fellow in the Zhang lab; Cathy Spangler, a graduate student in the McGinty lab; and Joshua Straus, PhD, assistant professor of biochemistry and biophysics and director of the CryoEM Core Facility at UNC-Chapel Hill. Other authors are Andrew Cesmat, a former technician at the UNC Eshelman School of Pharmacy who is currently a medical student at UNC, and Pengda Liu, PhD, assistant professor in the UNC Department of Biochemistry and Biophysics and UNC Lineberger member.
Funding for this research was provided through grants from the National Institutes of Health, a UNC School of Medicine Jefferson Pilot Fellowship to Qi Zhang, and Searle Scholars and Pew-Stewart Scholars awards to Robert McGinty. The Cryo-Electron Microscopy Facility, which is partially supported by the NIH, is part of the Molecular Microscopy Consortium of the University of North Carolina at Chapel Hill, Duke University, and the National Institute of Environmental Health Sciences.
–Written by Mark Derewicz
